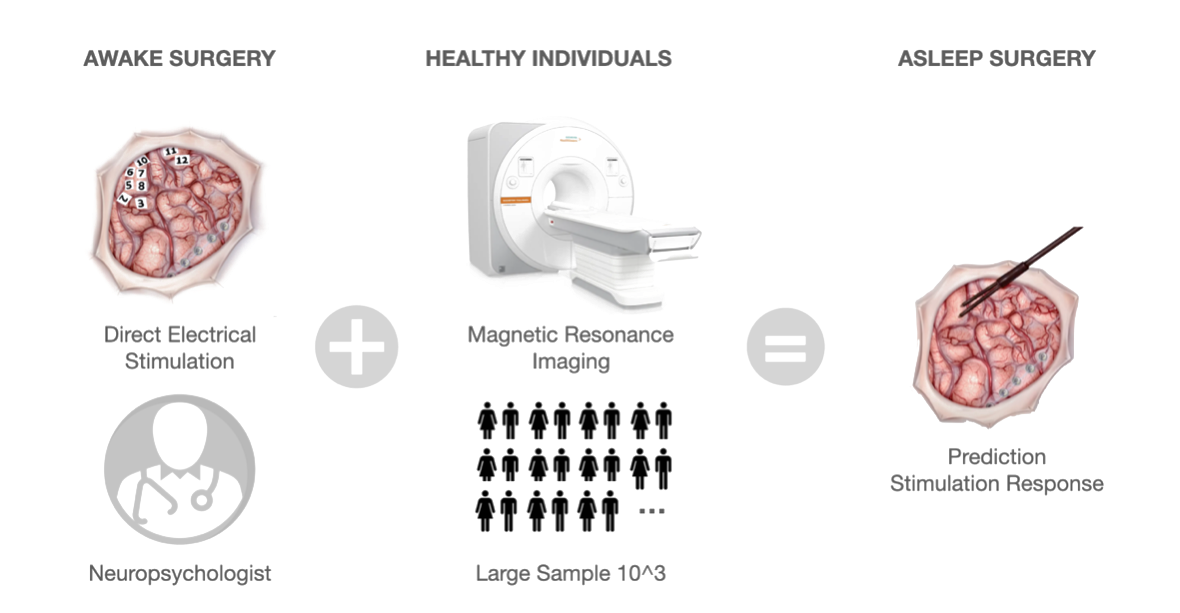
Direct electrical stimulation (DES) during cognitive and neurological monitoring is currently considered the most reliable brain mapping procedure for identifying brain regions critical for functional preservation. However, DES is unable to recover functional brain networks. By integrating DES with non-invasive functional connectome mapping, we unveiled the causal macroscale substrate subtending critical cognitive functions. Possible applications range from neurosurgery to psychiatry and neurology.
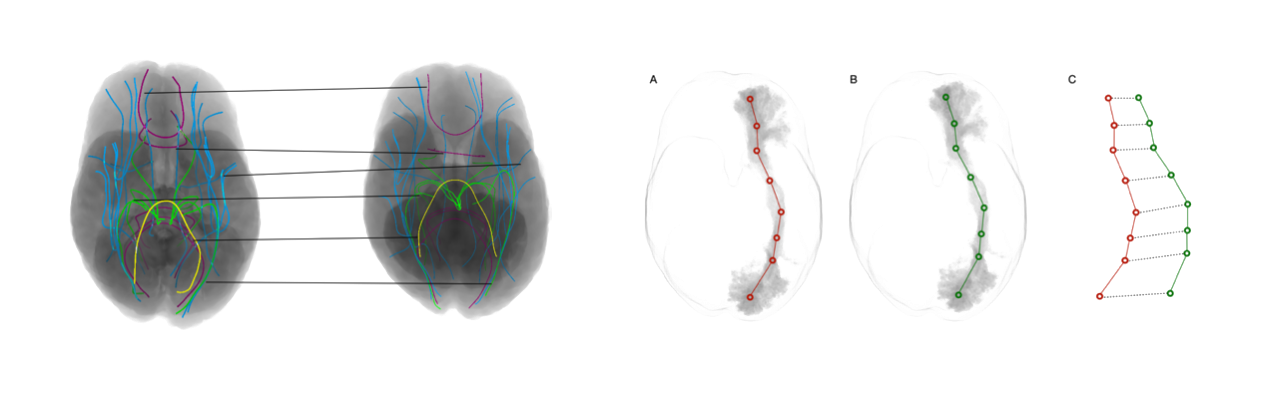
White matter pathways are encoded as tractogram data, representing 3D fiber pathways in the brain. Aligning tractograms is crucial for analyzing white matter across different subjects or mapping to a template. Current alignment methods rely on volumetric registration, which lacks fiber-specific orientation, leading to suboptimal results. This work proposes a Geometric Deep Learning solution to improve tractogram alignment by exploiting fiber correspondence, enabling supervised learning for more accurate alignment.
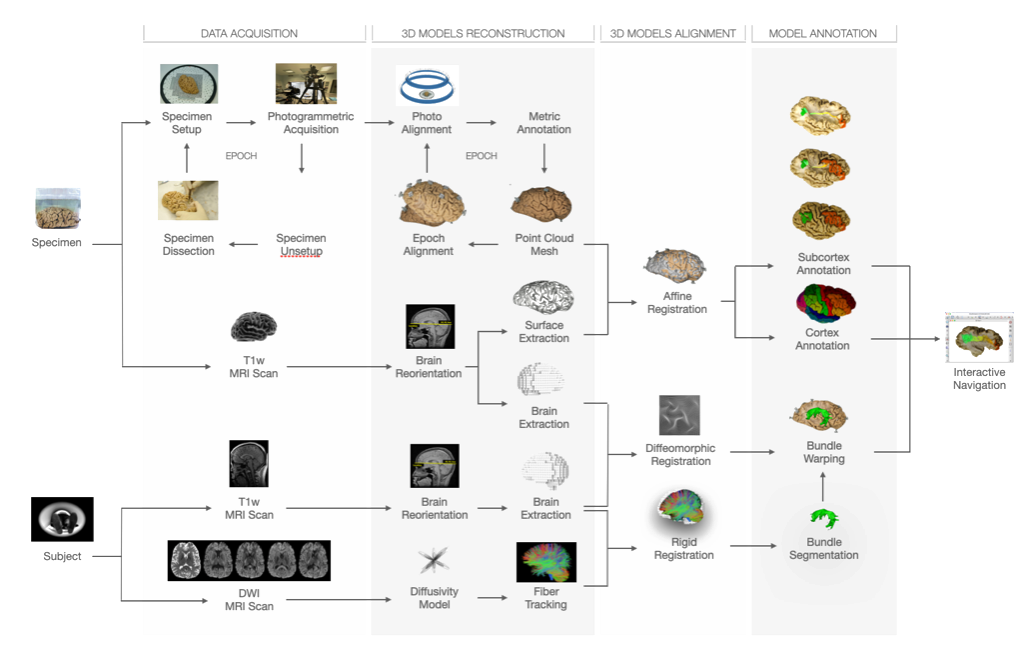
Photogrammetry was applied to cortex-sparing Klingler microdissection of the human brain, enabling accurate 3D reproduction of white matter neuroanatomy for validating tractography data. However, limited access to post-mortem brains and the complexity of dissection hindered generalization. To overcome these limitations, the BraDiPho (Brain Dissection Photogrammetry) framework was developed, offering a user-friendly platform to explore both microdissection and tractography data within the same environment.
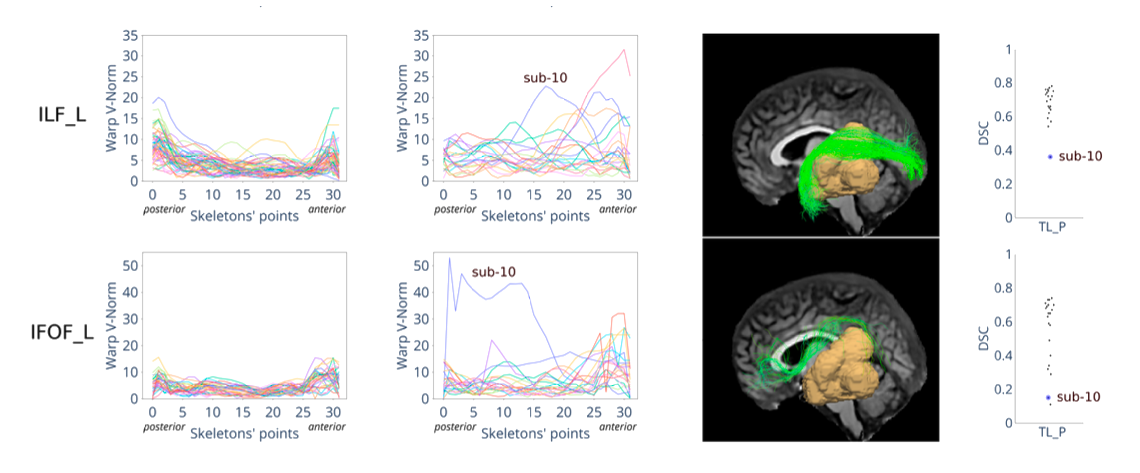
This project focuses on developing methods to segment white matter bundles in glioma patients automatically. This task is relevant to plan glioma resection surgery and reduces the risk of damage to the white matter bundles. Large public reference datasets are available for healthy populations, but not for glioma patients. We addressed this problem by using transfer learning from healthy population to glioma and we proved it to be successful in improving segmentation performance in glioma patients.
A tractogram is a virtual representation of brain white matter, consisting of millions of 3D polylines that approximate axonal pathways. Despite their accuracy in tasks like presurgical planning and brain disorder investigation, many tractogram fibers are artifacts. Verifyber addresses this by filtering non-plausible fibers using anatomical knowledge, rather than signal reconstruction or topology. Utilizing sequence Edge Convolution (sEC), Verifyber classifies fibers with >95% accuracy in under a minute for 1M fibers.
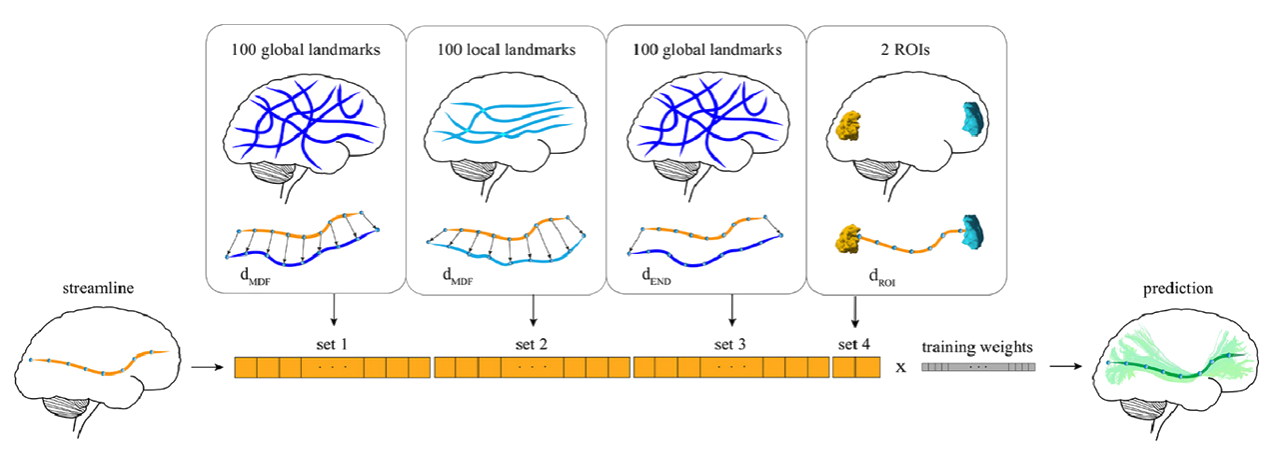
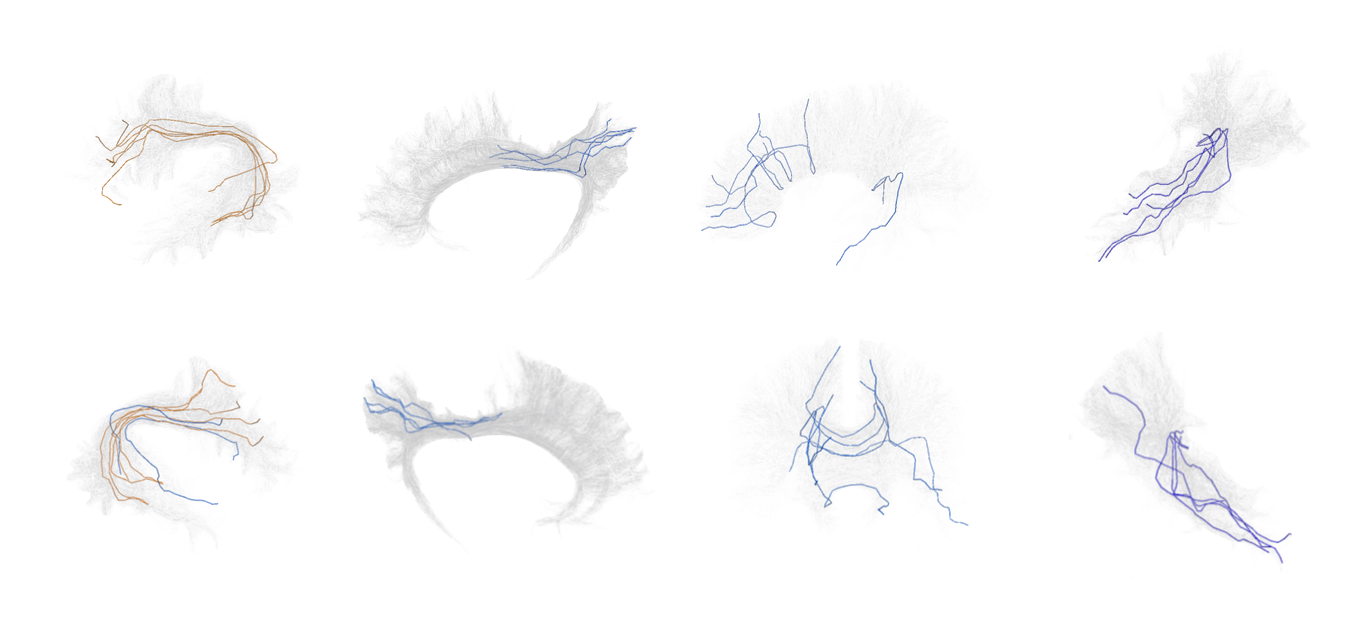
Classifyber is an AI-based method for automatic and accurate white matter bundle segmentation from tractography data, designed to assist in both research and clinical applications. It approaches segmentation as a supervised learning problem, using expert-based example bundles to learn segmentation rules. Classifyber extracts bundles by combining atlases, connectivity patterns, and fiber geometry. It outperforms state-of-the-art methods in accuracy, is fast, and aids in tasks like surgical planning and white matter assessment.
















