The functional and structural network maps aligned in MNI152 standard brain and derived from the integration of intracranial stimulation during awake surgery with normative structural and functional network mapping. The functional categories assessed during awake surgery were semantics, phonology, and speech articulation.
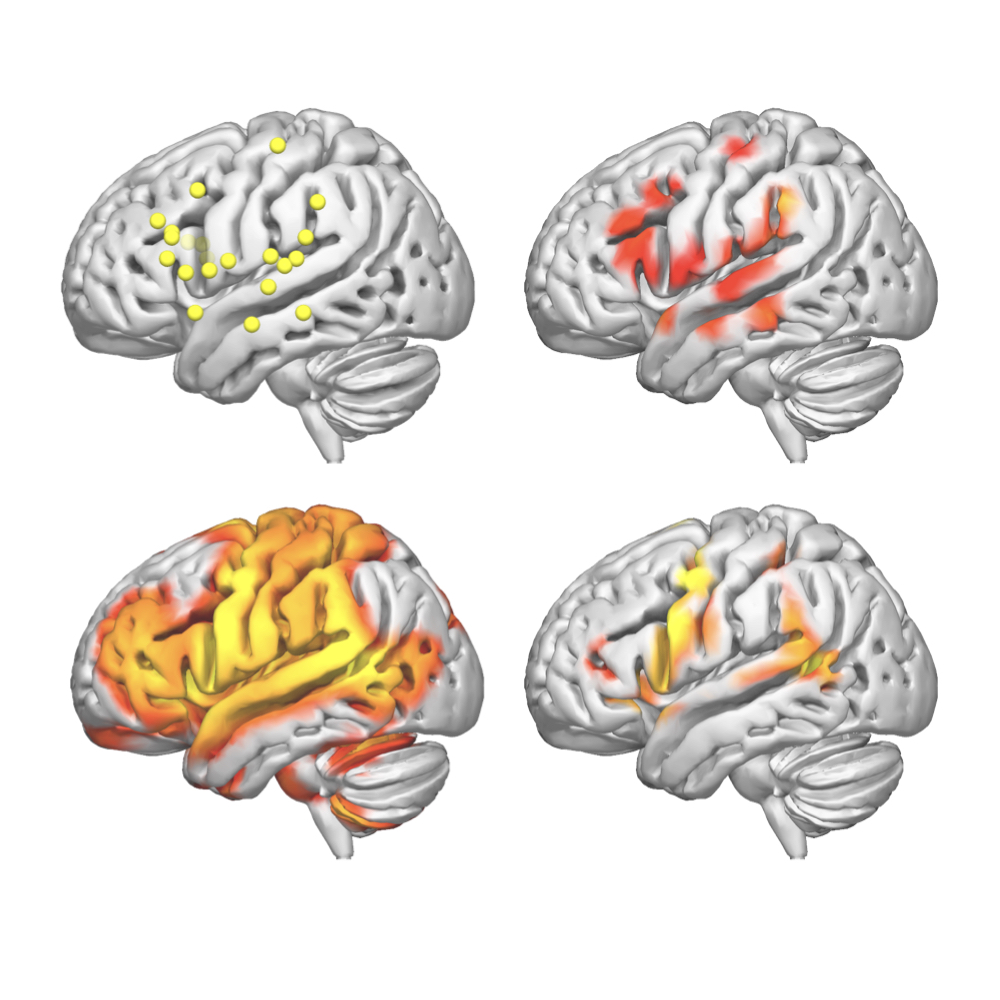
A collection of 2906 cortical and 1231 subcortical DES points derived from 592 low (WHO grade II) and 20 high grade (WHO grade III) glioma patients to provide a multimodal mapping of the networks underlying 12 functional domains. The 12 functional categories assessed during DES mapping were: verbal anomia, semantic and phonological paraphasia, non-verbal semantic association, motor and sensory responses, mentalizing, movement arrest, spatial perception, speech arrest, verbal apraxia and visual.
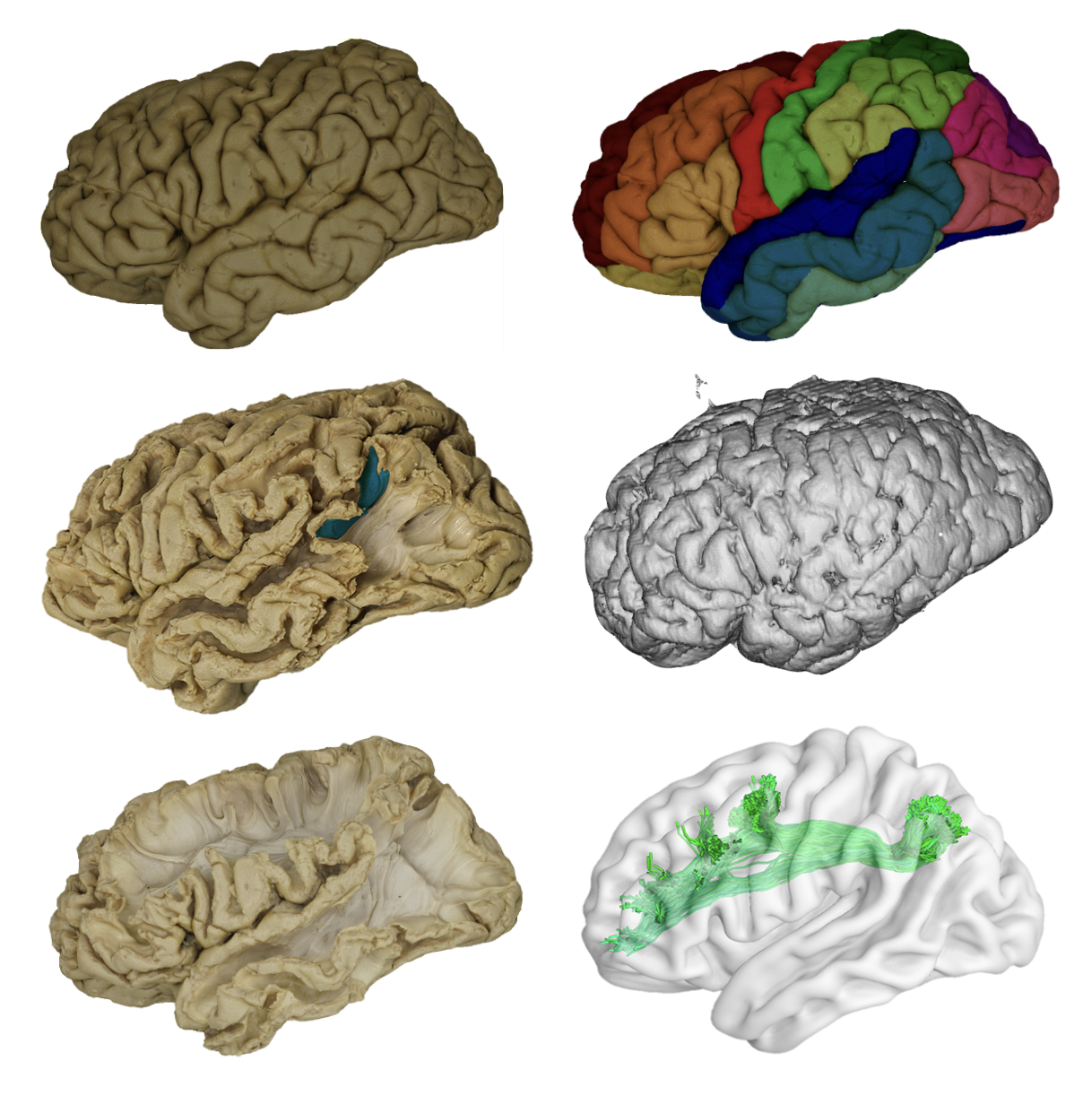
A collection of 3Dmodels of 12 ex-vivo specimen (six left and six right hemispheres), each demonstrating the progression of cortex-sparing Klingler dissection across 7 to 12 epochs per model. Each dissection layer is registered to a mesh model derived from the T1 MRI of the intact specimen. A series of twelve reference atlases registered within the radiological space of each BraDiPho model and each epoch. It comprises myelo- and cytoarchitectonic microstructural, functional and tractography-based human brain atlases.
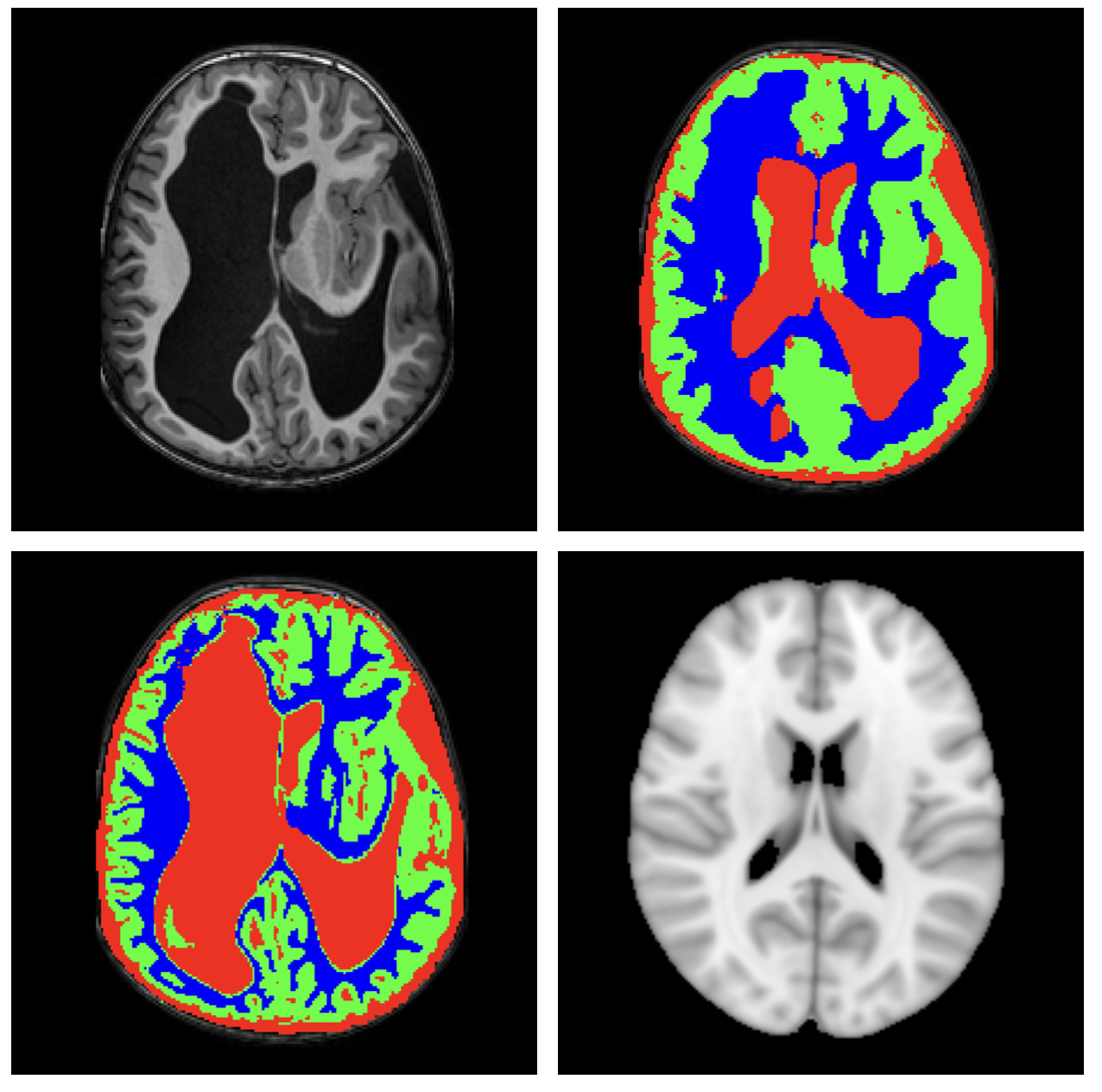
The dataset is composed of 918 healthy individuals drawn from three open datasets with T1-w images: NIH-Ped, C-Mind, Medea-Ped. The T1w images with distortions are organized into four different categories depending on the etiology: Agenesis of the Corpus Callosum (ACC), Posterior Fossa Malformations (PFM), Malformations of Cortical Development (MCDs), and severe distortion brains related to complex malformations or lesions (HD). All the T1w images are distributed with the annotation of brain tissues.
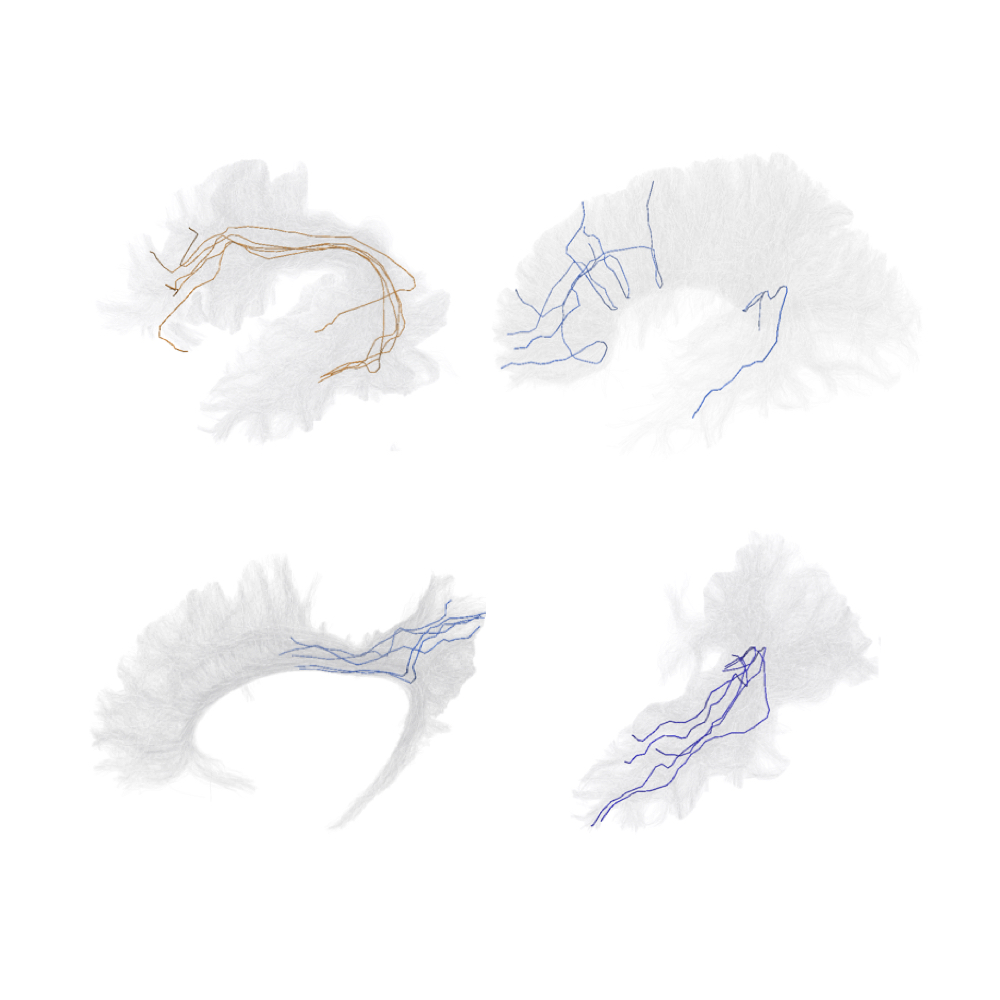
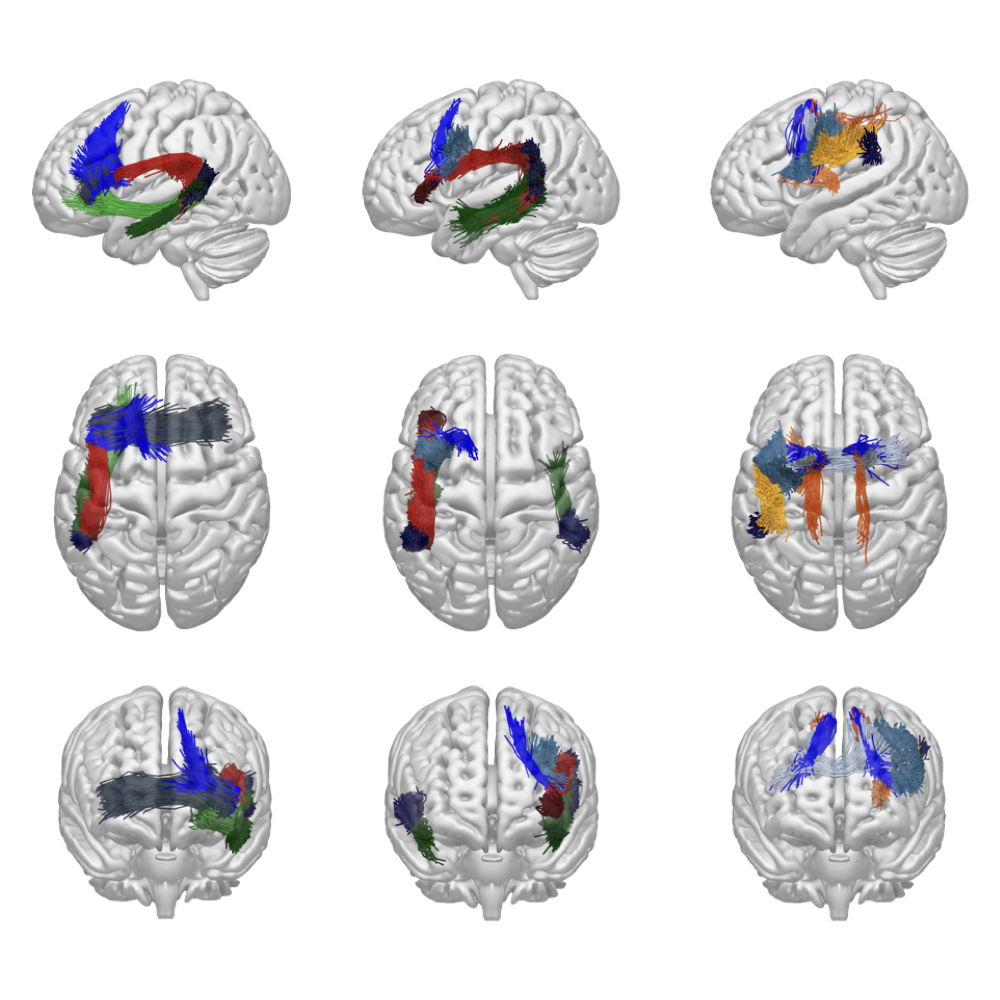
The dataset is composed of 20 individuals randomly selected from HCP repository. The processing pipeline carried out the estimation of the diffusivity model using the Constrained Spherical Deconvolution (CSD), and the fiber tracking using the algorithm for Particle Filtering Anatomically Constrained Tractography (PF-ACT). Tractograms were normalized to the same space via non-linear co-registration to the MNI152 standard brain. The binary labelling was performed using Extractor (Petit et al., 2022), a rule-based method that implements an exclusive policy to elicit the notion of anatomical plausibility.